fishing for electrons with thioredoxin domains
The QSOX family of disulfide bond-generating enzymes are ancient fusions of one or two thioredoxin domains linked to a helix-rich domain and then to an ERV domain. Algae, plants and protists have a single complete thioredoxin domain (blue bar) with a redox-active CxxC disulfide (Panel A). Metazoans contain an additional redox-inactive thioredoxin domain of uncertain function (pale blue; Panel B). A series of enzymological studies, along with the structure of the HRR-ERV pseudo-dimer, allowed us to construct a model for the flow of reducing equivalents between C1-C2 and C3-C4 CxxC motifs and the FAD cofactor (Panel C).
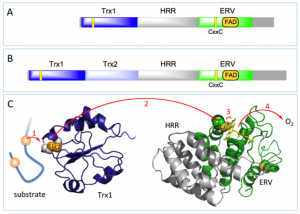 This model was cobbled together with fragments of structure, but until recently the structure of a full length QSOX was unavailable. Now Deborah Fass and her coworkers report the first crystal structures of protist and mammalian representatives of the QSOX family.
This model was cobbled together with fragments of structure, but until recently the structure of a full length QSOX was unavailable. Now Deborah Fass and her coworkers report the first crystal structures of protist and mammalian representatives of the QSOX family.
They first solved the structure of QSOX from the human parasite Trypanosoma brucei in both open and closed forms.
At the left is an open form of the enzyme about to receive reducing equivalents from a protein substrate. Once the C1-C2 disulfide on the thioredoxin domain of QSOX is reduced it must form a C1-C3 mixed disulfide with the ERV domain – and this entails a major conformation change. Fass and coworkers were able to catch the Trx domain in the act of passing electrons to the ERV domain by replacing the cysteines at the C2 and C4 positions with serine residues and then allowing the interdomain C1-C3 disulfide to form. The right panel is this closed conformation, in which the thioredoxin domain is pinned in place against the HRR-ERV domains. In the wild-type protein this intermediate is short-lived, and resolves with transfer of the pair of electrons to the flavin and the eventual reduction of oxygen. That frees up the tethered thioredoxin domain to start fishing for the next pair of electrons from QSOX substrates.
The stratagem used by Fass and coworkers to trap the closed conformation of the parasite enzyme was also used to get crystals of mouse QSOX1 [PubMed]. They show that the second mysterious thioredoxin domain (Trx2; light blue) is readily accommodated in the closed structure without interfering with the docking of the redox-active (Trx1) domain to the HRR-ERV domains. The Trx1-Trx2 half appears to move as a unit, and is again tethered to the HRR-ERV domains by a flexible and disordered linker.
These exciting new structures provide key insight into these promiscuous and proficient catalysts of protein disulfide bond formation. They help rationalize the enzymological data our groups have collected since the mid 1990’s and provide an essential structural context for the biological studies to follow.
Addendum
The Fass lab website is here.
The eagle-eyed will notice that the Thorpe group uses a different coloring system to the Fass group for the domains of QSOX – we color the Trx and Erv domains in blues and greens – whereas they reverse these colors!
There is a third absolutely conserved CxxC (C5-C6) motif in QSOXs that is shown at the bottom right of these crystal structures. Its role remains enigmatic.
The structure of the HRR-ERV pseudo-dimer of human QSOX1 was first reported here.
PDB Coordinates for Full length QSOXs
Trypanosoma brucei QSOX: open form 3QCP
Trypanosoma brucei QSOX: closed form3QD9
Mouse QSOX1: closed form 3T58
The crystal structures were displayed using PYMOL.


