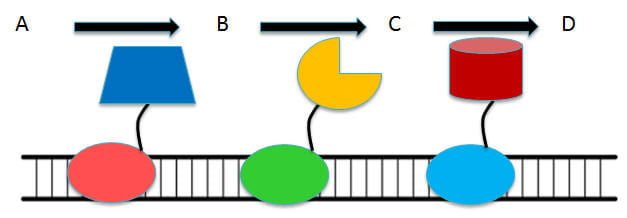
Cellular metabolism is capable of highly specific and efficient chemical synthesis at mild temperatures and pressures far beyond the capability of most synthetic chemical routes. Although pathway engineering can be used to further improve the range of compounds that could be synthesized, achieving commercially viable productivity remains challenging. An emerging strategy to combat these issues is to organize pathway enzymes into a sequential multi-enzyme complex in order to improve the overall pathway flux, to minimize cross-reactions, and to provide kinetic driving forces that redirect the carbon flux through essentially reversible steps. Although synthetic biologists have taken a more modular approach using biomolecular scaffolds to co-localize target enzymes, the current approaches either lack the required binding affinity or flexibility in tuning the cascade assembly. To address these shortcomings while providing a highly modular and flexible platform for assembling in vivo enzyme cascades, we propose here a new and potent approach that enables specific and high-affinity binding to DNA scaffolds using a special version of the CRISPR/Cas9 system. By combining the ability to provide site-specific, dCas9-guided enzyme assembly and the ability to provide disassembly by controlled dCas9 degradation, the overall goal is to develop a transformative framework to create highly efficient and dynamic enzyme cascades suitable for many synthetic-biology and metabolic engineering applications.
Project 2. Repurposing the CRISPR-Cas9 system for dynamic control of cellular metabolism (NSF) – with Prof. Papoutsakis